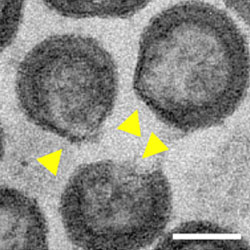
2023 Research Highlights
B-HIVE Member Presentations at the 2023 Retroviral Meeting at Cold Spring Harbor
Quantitative full-length sequencing of the HIV-1 transcriptome in primary cell derived resting CD4+ T-cells and T-cell subsets
Christian M. Gallardo, Nadejda Beliakova-Bethell, Bruce E. Torbett.
HIV-1 uncoating in the nucleus progresses through a distinct step of small defect formation in the capsid lattice prior to terminal uncoating
Levi Gifford, Gregory B. Melikian.
Mutations outside integrase lead to high-level resistance to integrase strand transfer inhibitors
Yuta Hikichi, Jonathan Grover, Walther Mothes, Eric Freed.
Structural insights into PPP2R5A degradation by HIV-1 Vif
Yingxia Hu, Krista A. Delviks-Frankenberry, Fidel Arizarga, Chunxiang Wu, Vinay Pathak, Yong Xiong.
Lenacapavir inhibits virion maturation by blocking formation of capsid pentamers
Szu-Wei Huang, Lorenzo Briganti, Guochao Wei, Arun Annamalai, Stephanie Bester, Reed Haney, Lauren Schope, Nikoloz Shkriabai, Ashwanth Francis, Daniel Adu-Ampratwum, James Fuchs, Alan Engelman, Mamuka Kvaratskhelia.
A critical role for the C-terminal mixed-charge domain of CPSF6 in nuclear HIV incursion and integration targeting
Sooin Jang, Hyun Jae Yu, Parmit K. Singh, Gregory J. Bedwell, Zachary M. Ingram, Satya P. Singh, AidanDarian Douglas, Zandrea Ambrose, Ashwanth C. Francis, Vineet N. KewalRamani, Alan N. Engelman.
Implications for integrase functional plasticity from the structure of the HIV-1 integrase tetramer
Tao Jing, Zelin Shan, Tung Dinh, Juliet Greenwood, Min Li, Terry Zhang, Bo Zhou, Sriram Aiyer, Robert Craigie, Alan N. Engelman, Mamuka Kvaratskhelia, Dmitry Lyumkis.
Lenacapavir disrupts HIV-1 core integrity while stabilizing the capsid lattice
Chenglei Li, Ryan C. Burdick, Sanath K. Janaka, Wei-Shau Hu, Vinay K. Pathak.
Intasomes assembled with full-length HIV-1 integrase and CTD facilitate structural studies by cryo-EM and reveal the role of the integrase C-terminal tail
Min Li, Xuemin Chen, Yanxiang Cui, Robert Craigie.
HIV-1 usurps transcription start site heterogeneity of host RNA polymerase II to maximize replication fitness
Olga Nikolaitchik, Saiful Islam, Jonathan Kitzrow, Alice Duchon, Zetao Cheng, Wei Shao, Maria Nikolaitchik, Mary Kearney, Frank Maldarelli, Karin Musier-Forsyth, Vinay K. Pathak, Wei-Shau Hu.
The role of LEDGF in transcription is intertwined with its function in HIV-1 integration
Rakesh Pathak, Parmit K. Singh, Hongen N. Zhang, Seyed Z. Rabi, Sahand Hormoz, Alan N. Engelman, Henry L. Levin.
Regulation of the timing of HIV-1 uncoating by nucleocapsid protein
Ioulia Rouzina, Helena Gien, Michael Morse, Micah McCauley, Jonathan Kitzrow, Karin Musier-Forsyth, Robert Gorelick, Mark Williams.
HIV-1 preintegration complex preferentially integrates the viral DNA into nucleosomes containing trimethylated histone 3-lysine36 modification and flanking linker DNA.
Nicklas Sapp, Nathaniel Burge, Khan Cox, Prem Prakash, Muthukumar Balasubramaniam, Devin Christenson, Min Li, Jared Linderberger, Mamuka Kvaratskhelia, Jui Pandhare, Robert Craigie, Michael Poirier, Chandravanu Dash.
Identification of host dependency factors that modulate nuclear import of HIV-1 cores
Rokeya Siddiqui, Sushila Kumari, Ryan C. Burdick, Gianluca Pegoraro, Wei-Shau Hu, Vinay K. Pathak.
CPSF6 licenses HIV-1 integration into Pol II-paused genes regulated by P-TEFb and U2 snRNP
Parmit K. Singh, Arun S. Annamalai, Mamuka Kvaratskhelia, Alan N. Engelman.
Live-cell and superresolution microscopy studies of HIV-1 restriction by MX2
Dariana Torres-Rivera, Gilberto Betancor, Michael H. Malim, Gregory B. Melikyan.
Antagonism of viral glycoproteins by guanylate binding protein 5
Hana Veler, Geraldine Vilmen, Abdul Waheed, Cheng Man Lun, Eric O. Freed.
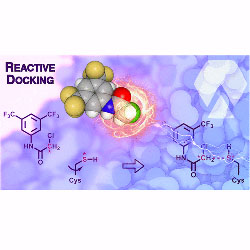
Reactive Docking: A computational method for high-throughput virtual screenings of reactive species
B-HIVE researchers develop a new method for discovering covalent inhibitors.
PubMed PMID: 37639635
Halo Library, a tool for rapid identification of ligand binding sites on proteins using crystallographic fragment screening
B-HIVE researchers show that halogenated compounds are an effective way to screen for new druggable sites on target proteins
PubMed PMID: 37115705
PubMedCentralPMCID: PMC10184123
Near-physiological in vitro assembly of 50S ribosomes involves parallel pathways
Ribosomes are a model system for profiling of complex ribonucleoprotein assembly.
PubMed PMID: 36864669
PubMedCentral PMCID: PMC10085682
Reversible phase separation of ESCRT protein ALIX through tyrosine phosphorylation
ALIX protein, which plays important roles in HIV budding, forms phase-separated condensates.
PubMed PMID: 37450591
PubMedCentral PMCID: PMC10348681

Multidisciplinary studies with mutated HIV-1 capsid proteins reveal structural mechanisms of lattice stabilization
By combining structural, biophysical and biochemical experiments, B-HIVE researchers reveal the role of key amino acids in HIV-1 capsid stability.
PubMed PMID: 37699872
PubMedCentral PMCID: PMC10497533

Critical mechanistic features of HIV-1 viral capsid assembly
B-HIVE researchers simulate the assembly of HIV capsid in the presence of IP6.
PubMed PMID: 36608139
PubMedCentral PMCID: PMC9821859

Structural insights into HIV-1 polyanion-dependent capsid lattice formation revealed by single particle cryo-EM
Cryo-EM structures of capsid in lattice assemblies reveal a critical role for inositol hexakisphosphate in the formation of pentamers.
PubMed PMID: 37094124
PubMedCentral PMCID: PMC10160977

Dynamics of upstream ESCRT organization at the HIV-1 budding site
Simulation is used to explore the role of cellular ESCRT proteins in the budding of HIV.
PubMed PMID: 37218128
PubMedCentral PMCID: PMC10397573
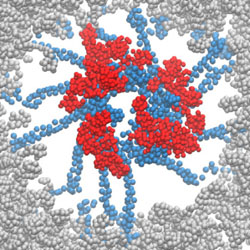
Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants
B_HIVE researchers study the mechanism of drug resistance in HIV-1 integrase and how some drugs evade resistance.
PubMed PMID: 37478179
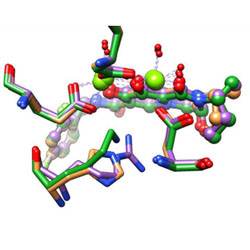
Targeting the HIV-1 and HBV Capsids, an EnCore
B-HIVE researchers discuss the targeting of capsid in antiviral therapy.
McFadden WM, Sarafianos SG. Viruses. 2023 Mar 31;15(4):896. doi: 10.3390/v15040896.
PubMed PMID: 37112877
PubMedCentral PMCID: PMC10146275
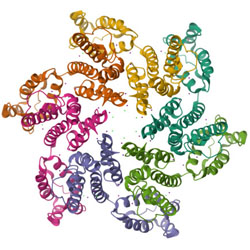
HIV-1 usurps transcription start site heterogeneity of host RNA polymerase II to maximize replication fitness
HIV-1 uses host-cell RNA polymerase II to create several slightly different viral RNAs with different specialized roles in viral replication.
PubMed PMID: 37252967
PubMedCentral PMCID: PMC10266039

A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase
RNA polymerases can successfully transcribe an expanded set of non-standard bases.
PubMed PMID: 38086811
PubMedCentral PMCID: PMC10716388

Hydrogen-deuterium exchange epitope mapping of glycosylated epitopes enabled by online immobilized glycosidase
Removing glycosylation can improve study of antibody binding epitopes by hydrogen-deuterium exchange and mass spectrometry.
PubMed PMID: 37379434

Assembly landscape for the bacterial large ribosomal subunit
The hierarchy of cooperative folding units is revealed in ribosome assembly.
PubMed PMID: 37633970
PubMedCentral PMCID: PMC10460392

The drug-induced interface that drives HIV-1 integrase hypermultimerization and loss of function
Atomic structures reveal the mechanism of integrase condensation by allosteric HIV-1 integrase inhibitors (ALLINIs).
PubMed PMID: 36744954
PubMedCentral PMCID: PMC9973045
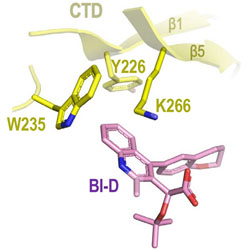
ViReMa: a virus recombination mapper of next-generation sequencing data characterizes diverse recombinant viral nucleic acids
An improved method is presented for identifying recombination events in long read sequences of viral genomes.
PubMed PMID: 36939008
PubMed Central PMCID: PMC10025937

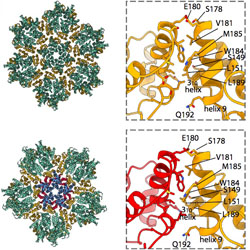
Two structural switches in HIV-1 capsid regulate capsid curvature and host factor binding
The inner mechanics of HIV-1 capsid are explored using single-particle cryoelectron microscopy.
PubMed PMID: 37040417
PubMedCentralPMCID: PMC10120081
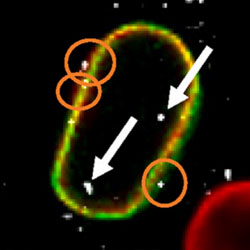
Label-free imaging of nuclear membrane for analysis of nuclear import of viral complexes
Individual HIV-1 replication complexes are visualized as they enter the cell nucleus.
PubMed PMID: 37875225
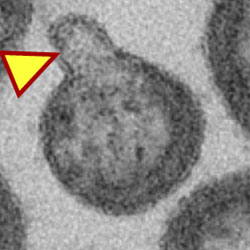
Neutral sphingomyelinase 2 is required for HIV-1 maturation
Neutral sphingomyelinase 2 plays important roles in HIV budding and maturation.
PubMed PMID: 37406093
PubMedCentral PMCID: PMC10334776
Inhibition of neutral sphingomyelinase 2 impairs HIV-1 envelope formation and substantially delays or eliminates viral rebound
Inhibitors of neutral sphingomyelinase 2 block HIV replication and kill HIV-infected cells.
PubMed PMID: 37406092
PubMedCentral PMCID: PMC10334757