2022 Research Highlights
B-HIVE Member Presentations at the 2022 Retroviral Meeting at Cold Spring Harbor
HIV-1 restriction by SERINC5 involves conformational changes in glycoprotein envelope that lead to loss of its function
Mariana Marin, Junghwa Kirschman, Gregory B. Melikyan
Manipulation of the central pore in capsomers reveals key insights into the role of IP6 in capsid function
Alex B. Kleinpeter, Donna L. Mallery, Nadine Renner, Leo C. James, Eric O. Freed
MX2 viral substrate breadth and inhibitory activity are regulated by protein phosphorylation
Gilberto Betancor, Madeleine Bangham, Steven Lynham, Jun K. Jeon, Kanisha Shah, Michael H. Malim
Prion-like low complexity regions enable avid virus-host interactions during HIV-1 infection
Guochao Wei, Naseer Iqbal, Ashwanth C. Francis, Arun S. Annamalai, Stephanie Bester, Parmit K. Singh, Szu-Wei Huang, Nikoloz Shkriabai, Reed Haney, Vineet N. KewalRamani, Alan N. Engelman, Gregory B. Melikyan, Francisco Asturias, Mamuka Kvaratskhelia
DDX3 and eIF4G1 regulate the kinetics of HIV-1 trafficking to nuclear speckles
Szu-Wei Huang, KyeongEun Lee, Hyun Jae Yu, Parmit K. Singh, Guochao Wei, Stephanie V. Rebensburg, Mamuka Kvaratskhelia, Alan Engelman, Vineet N. KewalRamani
Reverse transcription of long viral DNA is required to trigger efficient HIV-1 uncoating
Ryan C. Burdick, Wei-Shau Hu, Vinay K. Pathak
HIV-1 nucleocapsid protein regulates double-stranded DNA compaction and capsid uncoating during reverse transcription
Ioulia Rouzina, Helena Gien, Michael Morse, Micah J. McCauley, Jonathan P. Kitzrow, Karin Musier-Forsyth, Robert J. Gorelick, Mark C. Williams
Lenacapavir potently impairs HIV-1 integration inside the nucleus without displacing CPSF6 from viral capsid
Arun S. Annamalai, Guochao Wei, Naseer Iqbal, Ashwanth C. Francis, Stephanie Bester, Nikoloz Shkriabai, Gregory B. Melikyan, Francisco Asturias, Mamuka Kvaratskhelia
Timely chromatin invasion during mitosis dictates PFV integration site selection
Floriane Lagadec, Parmit K. Singh, Erik Serrao, Dirk Lindemann, Alan N. Engelman, Peter Cherepanov, Vincent Parissi, Paul Lesbats
Understanding the resistance profile of the second-generation INSTIs
Steven Smith, Min Li, Dario Passos, Xue Zhi Zhao, Robert Craigie, Dmitry Lyumkis, Terrence R. Burke, Jr, Stephen H. Hughes
Efforts toward deciphering mechanisms of resistance to integrase strand transfer inhibitors
Min Li, Dario O. Passos, Avik Biswas, Allan Haldane, Indrani Choudhuri, Zhaoyang Li, Nanjie Deng, Steven J. Smith, Xue-Zhi Zhao, Terrence R. Burke, Stephen H. Hughes, Ronald Levy, Robert Craigie, Dmitry Lyumkis
Simultaneous sequencing of full-length host cell and HIV-1 transcripts without PCR amplification
Christian M. Gallardo, Anh-Viet T. Nguyen, Andrew L. Routh, Bruce E. Torbett
Elements in the 5’ untranslated region of viral RNA important for Gag recognition
Zetao Cheng, Olga A. Nikolaitchik, Alice Duchon, Jonathan M. Rawson, Vinay K. Pathak, Wei-Shau Hu
Emergence of compensatory mutations reveal the importance of electrostatic interactions between HIV-1 integrase and genomic RNA
Christian Shema Mugisha, Tung Dinh, Kasyap Tenneti, Jenna E. Eschbach, Keanu Davis, Robert Gifford, Mamuka Kvaratskhelia, Sebla B. Kutluay
Inhibition of neutral sphingomyelinase 2 blocks HIV-1 maturation
Abdul A. Waheed, Seung Wan Yoo, Ferri Soheilian, Eva Agostino, Lwar Naing, Pragney Deme, Yanan Zhu, Yun Song, Peijun Zhang, Barbara Slusher, Norman Haughey, Eric Freed
Structures of immature HTLV-1 assemblies reveal a novel CA arrangement in deltaretrovirus particles
Martin Obr, Andreas Thader, Darya Chernikova, Gergely Pinke, Robert Dick, Florian Schur
High resolution structure determination of the mature HIV capsid lattice using cryo-ET and cryo-EM
Carolyn M. Highland, Aaron Tan, John A G. Briggs, Robert A. Dick
HTLV-1 Gag purification, cellular interactome, and in vitro chaperone activity
Yu-Ci Syu, Joshua Hatterschide, Yingke Tang, Jacob Al-Saleem, Amanda R. Panfil, Patrick L. Green, Karin Musier-Forsyth
Structural details of HIV maturation inhibitor binding determined by cryo-EM
Savannah G. Brancato, Robert A. Dick
Drug resistance and hypersensitivity of various HIV-1 subtypes to Islatravir
Maria E. Cilento, Philip R. Tedbury, Obiaara B. Ukah, Eiichi N. Kodama, Hiroaki Mitsuya, Michael A. Parniak, Karen A. Kirby, Stefan G. Sarafianos
Real-time imaging of HIV-1 protease activation in single newly assembled virions
Yinglin Li, Yen-Cheng Chen, Mariana Marin, Ashwanth C. Francis, Gregory B. Melikian
Immune reconstitution inflammatory syndrome drives emergence of HIV drug-resistance from multiple anatomic compartments
Camille Lange, Andrea Lisco, Christina Gavegnano, Maura Manion, Mary-Elizabeth Zipparo, Lindsey Adams, Frank Maldarelli, Irini Sereti
Immune correlates of cell-associated HIV RNA levels in infected individuals undergoing long-term antiretroviral therapy
Chuen-Yen Lau, Thuy Nguyen, Matthew Adan, Robert Gorelick, Jeannette Higgins, Jessica Earhart, Ariana Savramis, Catherine Rehm, Robin Dewar, Deborah McMahon, Anuradha Ganesan, Brian Luke, Frank Maldarelli
B-HIVE Member Presentations at the Twenty-Sixth West Coast Retrovirus Meeting
Session I: “The Power of Deep Sequencing”
Quantitative Full-length Sequencing of the HIV-1 Transcriptome in Quiescent CD4+ T-cells
Christian M. Gallardo, Nadejda Beliakova-Bethell, Bruce E. Torbett
Session IV: “Screens, Transcription & Latency”
KEYNOTE: Bruce E. Torbett, Seattle Children’s Research Institute and University of Washington
Proteomic Mapping of HIV-1 Gag-Host Interactions during Gag Assembly
Shentian Zhuang, Jade Wolff, Tien Le, Cade Ito, Mehrdad Alirezaei, Catherina Scharager-Tapia, Sung Kyu Park, John R. Yates III, George Tsaprailis, and Bruce E. Torbett
Poster Presentations
William McFadden, C. Highland, S. Brancato, A. Emanuelli, K. Kirby, Z. Wang, R. Dick, S. Sarafianos, Emory University, “HIV-1 Capsid Assemblies are Damaged by a Highly Potent, Caspid-Targeting Antiretroviral”
Alexa A. Snyder, MacQuillian J., Kaufman I., Song K., Du H., Kirby K.A., Michailidis E, Sarafianos S.G., Emory University, “Hypersensitivity of Non-nucleoside Resistant HIV-1 to Islatravir”
Shiyi Wang, Christian M. Gallardo, Davey M. Smith, James I. Mullins, Bruce E. Torbett, Seattle Children’s Research Institute, “Identification of HIV-1 Compensatory Mutational Networks Arising During Antiretroviral Therapy and the Contribution of Mutations to Drug Resistance Development and Improved Fitness”
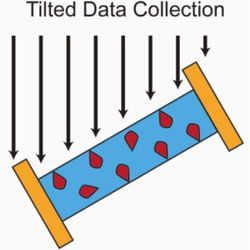
Single-particle cryo-EM data collection with stage tilt using Leginon
A protocol is presented to improve single particle cryo-EM structure determination by tilting specimens while collecting data.
PubMed PMID: 35848829
PubMedCentral PMCID: PMC9733953
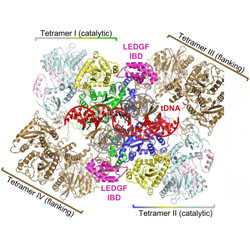
Multivalent interactions essential for lentiviral integrase function
Integrases form multi-subunit intasomes with DNA that may be important for targeting integration to active genes.
PubMed PMID: 35504909
PubMedCentral PMCID: PMC9065133
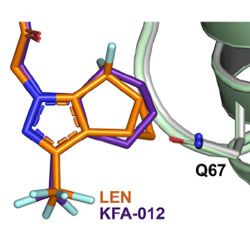
Structural and mechanistic bases of viral resistance to HIV-1 capsid inhibitor Lenacapavir
Structures of two important resistant mutants of HIV-1 capsid allowed the design of a more effective inhibitor.
PubMed PMID: 36190128
PubMedCentral PMCID: PMC9600929
MX2 viral substrate breadth and inhibitory activity are regulated by protein phosphorylation
Phosphorylation sites are identified on MX2 protein that increase HIV-1 inhibition and antiviral activity against mutant viruses.
PubMed PMID: 35880880
PubMedCentral PMCID: PMC9426416
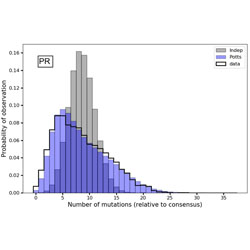
Limits to detecting epistasis in the fitness landscape of HIV
Interactions between mutations affect viral fitness, but may be difficult to observe experimentally.
PubMed PMID: 35041711
PubMedCentral PMCID: PMC8765623
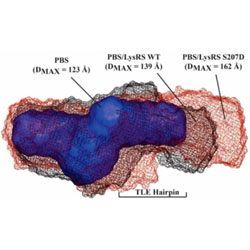
Phosphomimetic S207D lysyl-tRNA synthetase binds HIV-1 5’UTR in an open conformation and increases RNA dynamics
Phosphorylated lysyl-tRNA synthetase may be important for delivering primer tRNAs to HIV-1 genomic RNA.
PubMed PMID: 35891536
PubMedCentral PMCID: PMC9315659
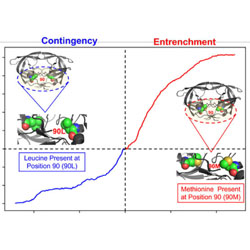
Contingency and entrenchment of drug-resistance mutations in HIV viral proteins
As HIV-1 mutates during antiretroviral therapy, drug resistance mutations are trapped by a combination of many accessory mutations.
PubMed PMID: 36493468
PubMedCentral PMCID: PMC9841799
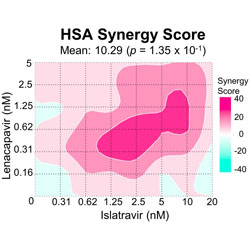
Drug interactions in lenacapavir-based long-acting antiviral combinations
Combinations of capsid inhibitors and reverse transcriptase inhibitors are explored for long-acting treatment of HIV-1.
PubMed PMID: 35746673
PubMedCentral PMCID: PMC9229705
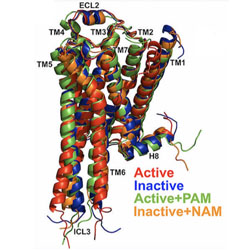
Unique features of different classes of G-protein-coupled receptors revealed from sequence coevolutionary and structural analysis
Covariation analysis of GPCR sequences is used to reveal sites involved in activation and allosteric regulation.
PubMed PMID: 34599827
PubMedCentral PMCID: PMC8738117
Multimodal functionalities of HIV-1 integrase
B-HIVE Center researchers review HIV-1 integrase mutants that cause defects in maturation.
Engelman AN, Kvaratskhelia M. Viruses. 2022 Apr 28;14(5):926. doi: 10.3390/v14050926.
PubMed PMID: 35632668
PubMedCentral PMCID: PMC9144474
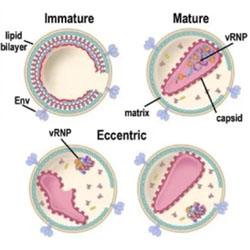
Localization and functions of native and eGFP-tagged capsid proteins in HIV-1 particles
Studies with eGFP-tagged capsid reveal that capsid remodeling is the predominant pathway for HIV-1 nuclear entry.
PubMed PMID: 35951676
PubMedCentral PMCID: PMC9426931
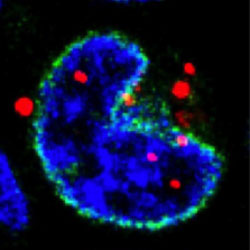
Selective ablation of 3′ RNA ends and processive RTs facilitate direct cDNA sequencing of full-length host cell and viral transcripts
A new sequencing method reveals alternative splicing of RNA within a host cell transcriptome.
PubMed PMID: 35736235
PubMedCentral PMCID: PMC9508845
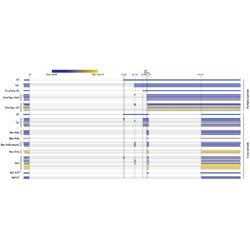
HIV-1 nucleocapsid protein binds double-stranded DNA in multiple modes to regulate compaction and capsid uncoating
Optical tweezers, fluorescence imaging, and atomic force microscopy are used to explore the interaction of HIV-1 nucleocapsid and DNA.
PubMed PMID: 35215829
PubMedCentral PMCID: PMC8879225

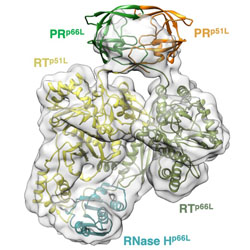
Cryo-EM structure of the HIV-1 Pol polyprotein provides insights into virion maturation
HIV-1 reverse transcriptase forms a similar dimer in the Pol polyprotein and in the mature form.
PubMed PMID: 35857464
PubMedCentral PMCID: PMC9258950
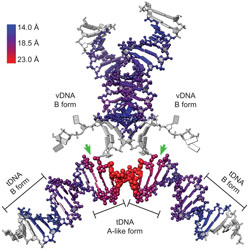
B-to-A transition in target DNA during retroviral integration
Retroviruses selectively integrate into regions of the genome where the host DNA can preferentially adopt the A-form.
PubMedPMID: 35947647
PubMedCentral PMCID: PMC9410886
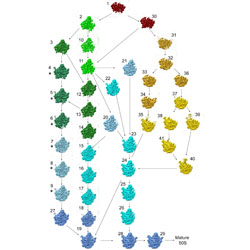
Quantitative mining of compositional heterogeneity in cryo-EM datasets of ribosome assembly intermediates
A workflow is presented that quantitatively mines cryo-EM data to isolate sets of structures with maximal diversity.
PubMed PMID: 34990602
PubMedCentral PMCID: PMC9891661
Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing
The structure, function and future uses of L-nucleoside inhibitors is reviewed.
PubMed PMID: 35218925
PubMedCentral PMCID: PMC9232924 (available on 2023-07-01)
TRIM5α restriction of HIV-1-N74D viruses in lymphocytes is caused by a loss of cyclophilin A protection
A mutation in HIV-1 capsid blocks binding to cyclophilin A, leading to reduced infectivity of the virus.
PubMed PMID: 35215956
PubMedCentral PMCID: PMC8879423
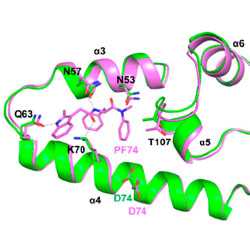
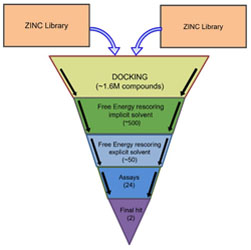
Structure-based virtual screening workflow to identify antivirals targeting HIV-1 capsid
A combination of computational docking and advanced free energy estimation is used to discover new HIV-1 inhibitors.
PubMed PMID: 35262811
PubMedCentral PMCID: PMC8904208
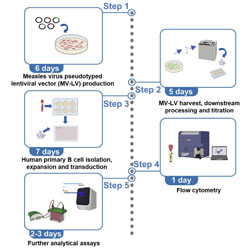
An optimized measles virus glycoprotein-pseudotyped lentiviral vector production system to promote efficient transduction of human primary B cells
A efficient protocol is presented to package complex transgene-containing lentiviral vectors.
PubMed PMID: 35284833
PubMedCentral PMCID: PMC8914380
Covariation of viral recombination with single nucleotide variants during virus evolution revealed by CoVaMa
Analysis of Flock House virus sequences reveals that recombination and point mutations of the genome are intimately linked.
PubMed PMID: 35018461
PubMedCentral PMCID: PMC9023271
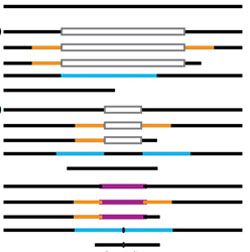
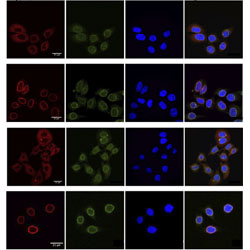
Prion-like low complexity regions enable avid virus-host interactions during HIV-1 infection
Several host cell proteins bind to HIV-1 capsids using unusual prion-like segments.
PubMed PMID: 36202818
PubMedCentral PMCID: PMC9537594
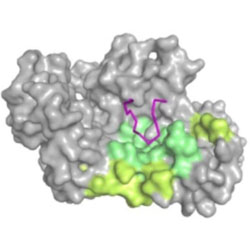
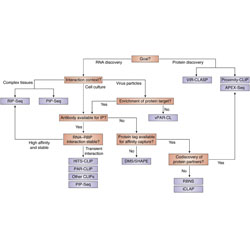
Next-generation sequencing: A new avenue to understand viral RNA-protein interactions
Recent advances in next-generation sequencing technologies have allowed the characterization of viral RNA-protein interactions.
PubMed PMID: 35413291
PubMedCentral PMCID: PMC8994257