2021 Research Highlights
HIVE Member Presentations at the 2021 Retroviral Meeting at Cold Spring Harbor
INTERACTIONS BETWEEN HIV-1 REVERSE TRANSCRIPTASE AND HUMAN APOBEC3G
Ryan L. Slack, Darja Pollpeter, Scott J. Novick, Seongmi Kim, Kamlendra Singh, Eddy Arnold, Patrick R. Griffin, Michael H. Malim, Stefan G. Sarafianos.
A NOVEL BIS(ARYL FLUOROSULFATE) COMPOUND COVALENTLY BINDS TO THE HIV-1 CAPSID PROTEIN
Mary C. Casey, Karen A. Kirby, Hua Wang, Theresa Tiefenbrunn Sample, Philip R. Tedbury, John R. Cappiello, Bruce E. Torbett, Arthur J. Olson, K Barry Sharpless, Stefan G. Sarafianos.
ADAPTATION OF CHIMERIC HIV—INSIGHTS INTO GAG-RNA INTERACTIONS THAT LEAD TO GENOME PACKAGING
Jonathan M. Rawson, Olga A. Nikolaitchik, Jennifer A. Yoo, Xayathed Somoulay, Vinay K. Pathak, Ryan L. Slack, Stefan G. Sarafianos, Wei-Shau Hu.
MULTIOMICS PROFILING OF HIV-INFECTED CELLS REVEALS UNIQUE CHROMATIN ‘SIGNATURE’ ASSOCIATED WITH ACTIVE INTEGRATED PROVIRUSES
Raven Shah, Christian M. Gallardo, Yoonhee H. Jung, Victor G. Corces, Bruce E. Torbett, Philip R. Tedbury, Stefan G. Sarafianos.
IDENTIFY HOST FACTORS INVOLVED IN GAG TRAFFICKING AND ASSEMBLY BY USING SPLIT-APEX2 SYSTEM COUPLED WITH QUANTITATIVE MS
Shentian Zhuang, Bruce Torbett
MRHAMER YIELDS HIGHLY ACCURATE SINGLE MOLECULE VIRAL SEQUENCES ENABLING ANALYSIS OF INTRA-HOST EVOLUTION
Christian M. Gallardo, Shiyi Wang, Daniel J. Montiel-Garcia, Susan J. Little, Davey M. Smith, Andrew L. Routh, Bruce E. Torbett.
SHAPE PROBING OF THE HIV-1 REVERSE TRANSCRIPTASE INITIATION COMPLEX REVEALS RNA FLEXIBILITY CHANGES ADJACENT TO THE PRIMERBINDING SITE
Chathuri Pathirage, William Cantara, Steven Tuske, Eddy Arnold, Karin Musier-Forsyth.
ROLE OF HIV-1 5’UTR RNA STRUCTURE AND CONFORMATIONAL DYNAMICS IN GENOME PACKAGING
Jonathan P. Kitzrow, Shuohui Lui, Shiqin Miao, Kevin Jamison, Dennis Bong, Michael Poirier, Karin Musier-Forsyth.
ANALYSIS OF HTLV-1 GAG INTERACTOME AND CHARACTERIZATION OF ITS CHAPERONE ACTIVITY
Yu-Ci Syu, Joshua Hatterschide, Yingke Tang, Jacob Al-Saleem, Amanda R. Panfil, Patrick L. Green, Karin Musier-Forsyth.
STRUCTURE OF HIV-1 POL POLYPROTEIN REVEALS REVERSE TRANSCRIPTASE P66/P51 HETERODIMER-LIKE CORE: IMPLICATIONS FOR PROTEASE ACTIVATION
Jerry J Harrison, Dario Oliveira Passos, Jessica Bruhn, Joseph Bauman, Lynda Tuberty, Jeffrey DeStefano, Dmitry Lyumkis, Eddy Arnold.
HIGH RESOLUTION MAPPING OF GENIC TARGETING PREFERENCES DURING HIV-1 INTEGRATION IN VITRO AND IN VIVO
Gregory J. Bedwell, Sooin Jang, Wen Li, Parmit K. Singh, Alan N. Engelman.
STRUCTURAL BASIS FOR THE EVOLUTION OF VIRAL RESISTANCE TO LONG-ACTING HIV-1 CAPSID INHIBITOR GS-6207
Stephanie M. Bester, Arun S. Annamalai, Daniel Adu-Ampratwum, Reed Haney, James R. Fuchs, Mamuka Kvaratskhelia.
PRE-MRNA SPLICING AND GENE DENSITY ARE SIGNIFICANT DETERMINANTS OF LENTIVIRAL INTEGRATION
Parmit K. Singh, Vidya Chivukula, Peter Cherepanov, Alan N. Engelman.
GFP-TAGGED HIV-1 CAPSID PROTEIN IS SELECTIVELY PACKAGED WITH VIRAL RIBONUCLEOPROTEIN COMPLEX INSIDE THE CONICAL SHELL OF INFECTIOUS VIRIONS
Ashwanth C Francis, Parmit K Singh, Jiong Shi, Anna Cereseto, Alan Engelman, Christopher R Aiken, Gregory B Melikyan
HIV-1 INTEGRATION FAVORS GENES ASSOCIATED WITH PROMOTER-PROXIMAL POL II PAUSING
Parmit K Singh, Arun Annamalai, Mamuka Kvaratkshelia, Alan N. Engelman
UNVEILING THE GENETIC FRAGILITY OF HIV-1 THROUGH DEEP LEARNING
Juan S. Rey, Wen Li, Hagan Beatson, Christian Lantz, Alexander Bryer, Alan N. Engelman, Juan R. Perilla.
HIV-1 PIC-MEDIATED INTEGRATION IS SELECTIVELY AFFECTED BY NUCLEOSOME STRUCTURE
Nicklas Sapp, Nathan Burge, Khan Cox, Muthukumar Balasubramaniam, Jui Pandhare, Min Li, Jared Lindenberger, Mamuka Kvaratskhelia, Robert Craigie, Michael Poirier, Chandravanu Dash
DDX3 AND EIF4G1 INFLUENCE HIV-1 TRAFFICKING TO NUCLEAR SPECKLES
Szu-Wei Huang, KyeongEun Lee, Hyun Jae Yu, Stephanie V. Rebensburg, Mamuka Kvaratskhelia, Vineet N. KewalRamani.
Evaluating local and directional resolution of cryo-EM density maps
Methods are presented for evaluating and improving cryo-EM maps.
Aiyer S, Zhang C, Baldwin PR, Lyumkis D. Methods Mol Biol. 2021;2215:161-187.
PubMed PMID: 33368004
Tools for visualizing and analyzing Fourier space sampling in Cryo-EM
Tools are presented for understanding how molecular orientation impacts structure determination by cryoelectron microscopy.
Baldwin PR, Lyumkis D. Prog Biophys Mol Biol. 2021 Mar;160:53-65.
PubMed PMID: 32645314
PubMedCentral PMCID: PMC7785567
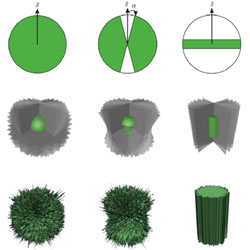
rigrag: high-resolution mapping of genic targeting preferences during HIV-1 integration in vitro and in vivo
A method is presented for identifying genes that are recurrently targeted by HIV integrase, and genes that are recurrently avoided.
Bedwell GJ, Jang S, Li W, Singh PK, Engelman AN. Nucleic Acids Res. 2021 Jul 21;49(13):7330-7346.
PubMed PMID: 34165568
PubMedCentral PMCID: PMC8287940
MX2-mediated innate immunity against HIV-1 is regulated by serine phosphorylation
Several cellular proteins are identified that interact with MX2 as part of its antiviral action.
PubMed PMID: 34282309
PubMedCentral PMCID: PMC7611661
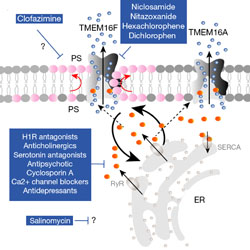
Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia
Three thousand approved drugs are screened for use in COVID-19 therapy.
PubMed PMID: 33827113
PubMedCentral PMCID: PMC7611055
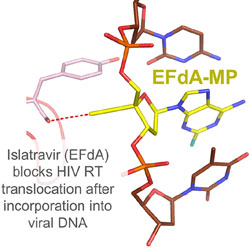
Avoiding drug resistance in HIV reverse transcriptase
HIVE Center researchers review the biology of HIV reverse transcriptase and discuss approaches to fight drug resistance.
Cilento ME, Kirby KA, Sarafianos SG. Chem Rev. 2021 Mar 24;121(6):3271-3296.
PubMed PMID: 33507067
PubMedCentral PMCID: PMC8149104
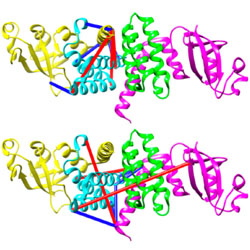
Revealing the structural plasticity of SARS-CoV-2 nsp7 and nsp8 using structural proteomics
Solution-based structural proteomics techniques reveal that two coronavirus non-structural proteins may form multiple oligomeric states.
PubMed PMID: 34121407
PubMedCentral PMCID: PMC8231661
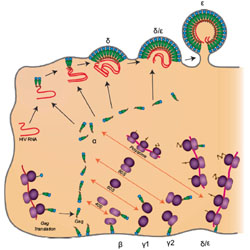
Discrimination between functional and non-functional cellular Gag complexes involved in HIV-1 assembly
HIV Gag protein bound to cellular ribosomes may acts a reservoir of Gag during assembly of new viruses
PubMed PMID: 33539875
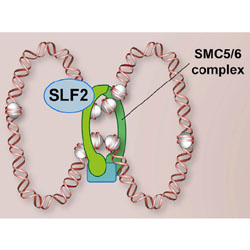
The SMC5/6 complex compacts and silences unintegrated HIV-1 DNA and is antagonized by Vpr
Researchers explore how HIV protein Vpr blocks the natural defenses against integration of the viral genome.
PubMed PMID: 33811831
PubMedCentral PMCID: PMC8118623
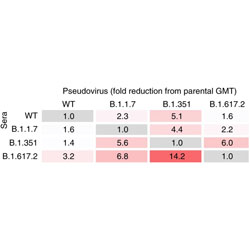
Neutralizing antibody activity in convalescent sera from infection in humans with SARS-CoV-2 and variants of concern
Individuals infected with different strains of SARS-CoV-2 produce cross-neutralizing antibodies, but they many not be as effective against divergent lineages of the virus.
PubMed PMID: 34654917
PubMedCentral PMCID: PMC8556155
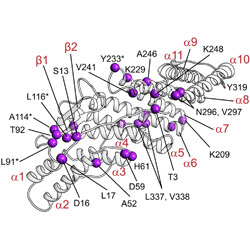
Mechanistic roles of tyrosine phosphorylation in reversible amyloids, autoinhibition, and endosomal membrane association of ALIX
An autoinhibition mechanism relocates ALIX to the cytosol and tyrosine phosphorylation plays many roles in its functions.
Elias RD, Ramaraju B, Deshmukh L. J Biol Chem. 2021 Nov;297(5):101328.
PubMed PMID: 34688656
PubMedCentral PMCID: PMC8577116
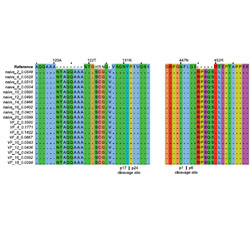
MrHAMER yields highly accurate single molecule viral sequences enabling analysis of intra-host evolution
A new nanopore-based method allows long-range sequencing of thousands of viral genomes from clinical samples.
PubMed PMID: 33849057
PubMedCentral PMCID: PMC8266615
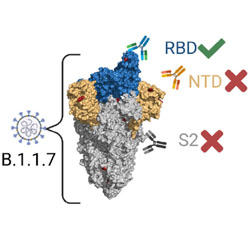
Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant
A panel of over a hundred antibodies are used to explore how SARS-CoV-2 evades neutralization by the immune system.
PubMed PMID: 33836142
PubMedCentral PMCID: PMC8015430
Mi3-GPU: MCMC-based Inverse Ising Inference on GPUs for protein covariation analysis
HIVE Center members improve methods for predicting interactions between amino acids based on observed mutational covariations.
Haldane A, Levy RM. Comput Phys Commun. 2021 Mar;260:107312.
PubMed PMID: 33716309
PubMedCentral PMCID: PMC7944406
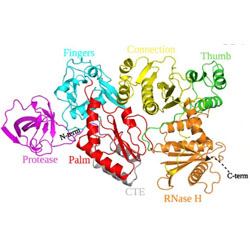
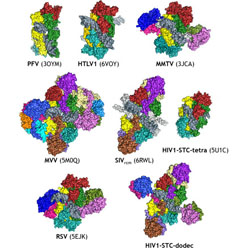
Crystal structure of a retroviral polyprotein: Prototype foamy virus protease-reverse transcriptase (PR-RT)
The structure of a monomeric precursor of PFV protease-reverse transcriptase is presented.
PubMed PMID: 34452360
PubMedCentral PMCID: PMC8402755
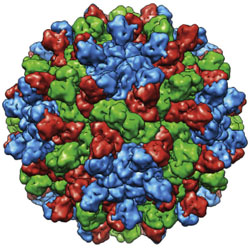
Icosahedral virus structures and the Protein Data Bank
The history of structural studies of icosahedral viruses is reviewed.
Johnson JE, Olson AJ. J Biol Chem. 2021 Jan-Jun;296:100554.
PubMed PMID: 33744290
PubMedCentral PMCID: PMC8081926
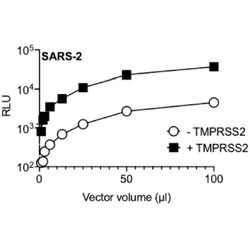
TMPRSS2 promotes SARS-CoV-2 evasion from NCOA7-mediated restriction
Evidence is found that SARS-CoV-2 enters cells by two different pathways as a way to avoid restriction.
PubMed PMID: 34807954
PubMedCentral PMCID: PMC8648102
Resilient SARS-CoV-2 diagnostics workflows including viral heat inactivation
Commercially-available methods for detection of SARS-CoV-2 nucleic acid are compared.
PubMed PMID: 34525109
PubMedCentral PMCID: PMC8443028
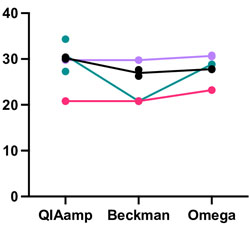
HIV-1 integrase binding to genomic RNA 5′-UTR induces local structural changes in vitro and in virio
HIVE Center researchers find that nucleocapsid and integrase bind differently to HIV genomic RNA
PubMed PMID: 34809662
PubMedCentral PMCID: PMC8609798
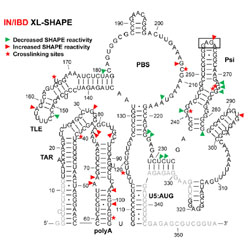
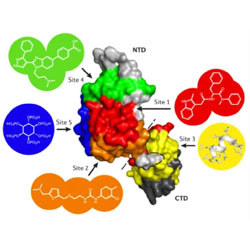
Rotten to the core: antivirals targeting the HIV-1 capsid core
The role of HIV-1 capsid in the viral life cycle and the mechanisms of capsid inhibitors are reviewed.
PubMed PMID: 34937567
PubMedCentral PMCID: PMC8693499
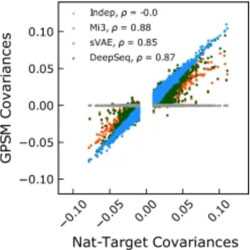
The generative capacity of probabilistic protein sequence models
Several methods for predicting evolutionary sequence covariation in viral populations are tested and compared.
PMID: 34728624
PMCID: PMC8563988
Selective packaging of HIV-1 RNA genome is guided by the stability of 5′ untranslated region polyA stem
The stability of the polyA stem guides selective packaging of different forms of the HIV-1 genomic RNA.
PubMed PMID: 34873042
PubMedCentral PMCID: PMC8685901
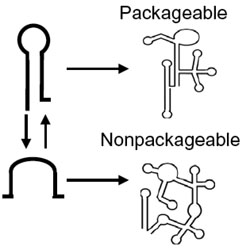
Retroviral integrase: Structure, mechanism, and inhibition
The function, structure, and inhibition of retroviral integrases are reviewed.
PubMed PMID: 34861940
PubMedCentral PMCID: PMC8732146
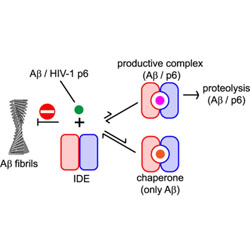
Quantitative NMR study of insulin-degrading enzyme using amyloid-β and HIV-1 p6 elucidates its chaperone activity
Insulin-degrading enzyme forms a single productive complex with HIV p6 and a more complex set of interactions with amyloids.
Ramaraju B, Nelson SL, Zheng W, Ghirlando R, Deshmukh L. Biochemistry. 2021 Aug 24;60(33):2519-2523.
PubMedCentral PubMed PMID: 34342986
Sec24C is an HIV-1 host dependency factor crucial for virus replication
Cellular protein Sec24C plays essential roles in the trafficking of HIV-1.
PubMed PMID: 33649557
PubMedCentral PMCID: PMC8012256
Integrase strand transfer inhibitors are effective anti-HIV drugs
HIV Center members and collaborators review the current stated of clinically-relevant integrase strand transfer inhibitors.
PubMed PMID: 33572956
PubMedCentral PMCID: PMC7912079
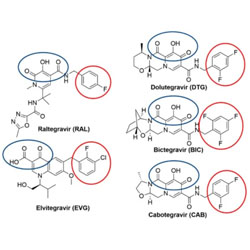
HIV-1 integrase inhibitors with modifications that affect their potencies against drug resistant integrase mutants
Based on structures of HIV-1 and SIV integrases, new integrase strand transfer inhibitors are designed and tested for their ability to block drug-resistant mutants of the enzyme.
PubMed PMID: 33686850
PubMedCentral PMCID: PMC8205226
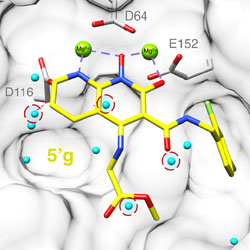
Molecular dynamics free energy simulations reveal the mechanism for the antiviral resistance of the M66I HIV-1 capsid mutation
The free energy cost of protein side chain reorganization is largely responsible for drug resistance of a capsid inhibitor.
Sun Q, Levy RM, Kirby KA, Wang Z, Sarafianos SG, Deng N. Viruses. 2021 May 15;13(5):920.
PMID: 34063519
PMCID: PMC8156065
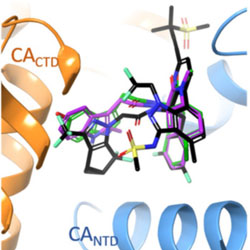
Computational design of small molecular modulators of protein-protein interactions with a novel thermodynamic cycle: Allosteric inhibitors of HIV-1 integrase
A new method for targeting protein-protein interfaces is applied to design of integrase inhibitors
Sun Q, Ramaswamy VSK, Levy R, Deng N. Protein Sci. 2021 Feb;30(2):438-447.
PubMed PMID: 33244804
PubMedCentral PMCID: PMC7784772
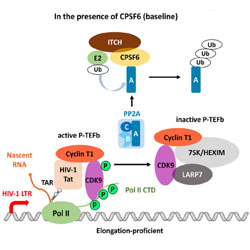
Cleavage and Polyadenylation Specificity Factor 6 is required for efficient HIV-1 latency reversal
The cellular protein CPSF6 plays a novel role in HIV-1 transcription and latency of the virus.
PubMed PMID: 34154414
Interactions of HIV-1 Capsid with host factors and their implications for developing novel therapeutics
The structure and function of HIV-1 capsid and its interactions with host factors and drugs are reviewed.
Zhuang S, Torbett BE. Viruses. 2021 Mar 5;13(3):417.
PubMed PMID: 33807824
PubMedCentral PMCID: PMC8001122