2016 Research Highlights
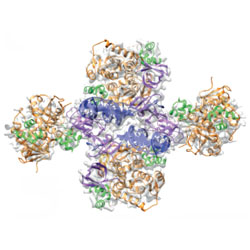
Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function
An unexpected octameric structure for a retroviral intasome is revealed using single-particle cryo-electron microscopy.
PubMed PMID: 26887496
PubMed Central PMCID: PMC4908968
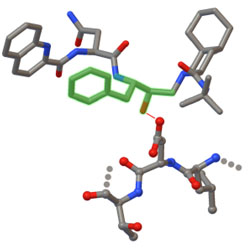
Rapid experimental SAD phasing and hot-spot identification with halogenated fragments
Small molecules with halogens are shown to be an effective new tool for solving the phase problem in x-ray crystallography.
Bauman JD, Harrison JJEK & Arnold E IUCrJ 3, 51-60 (2016)
PubMed PMID: 26870381
PubMedCentral PMCID: PMC4704079

Fragment-based analysis of ligand dockings improves classification of actives
Results from virtual screening are analyzed to improve ranking of compounds for discovery of new inhibitors.
Belew RK, Forli S, Goodsell DS, O’Donnell TJ & Olson AJ J Chem. Inf. Model. 56, 1597-1607 (2016)
PubMed PMID: 27384036
PubMedCentral PMCID: 5023760
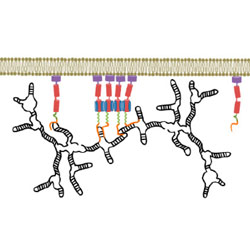
On the selective packaging of genomic RNA by HIV-1
HIVE Center collaborators review the way that HIV-1 packages its genomic RNA
Comas-Garcia M, Davis SR & Rein A Viruses 8, 246 (2016)
PubMed PMID: 27626441
PubMedCentral PMCID: PMC5035960
Conformational states of HIV-1 reverse transcriptase for nucleotide incorporation vs pyrophosphorolysis–binding of foscarnet
A DNA aptamer is used to trap complexes of reverse transcriptase in a new structural state, providing insights for developing new inhibitors.
PubMed PMID: 27192549
PubMedCentral PMCID: PMC4992415

Dimerization of the SP1 region of HIV-1 Gag induces a helical conformation and association into helical bundles: Implications for particle assembly
HIVE Center collaborators explore the central role of the SP1 spacer of Gag in assembly of HIV-1.
PubMed PMID: 26637452
PubMedCenter PMCID: PMC4733982

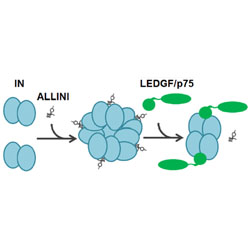
Allosteric HIV-1 integrase inhibitors promote aberrant protein multimerization by directly mediating inter-subunit interactions: structure and thermodynamic modeling studies
ALLINIs stabilize the interaction between integrase catalytic domains and C-terminal domains, causing aggregation that impares virus maturation.
PubMed PMID: 27503276
PMCID: PMC5079246

The competitive interplay between allosteric HIV-1 integrase inhibitor BI/D and LEDGF/p75 during the early stage of HIV-1 replication adversely affects inhibitor potency
Binding of the cellular protein LEDGF/p17 blocks the multimerization of HIV-1 integrase by an ALLINI, reducing the inhibitor’s antiviral activity.
PubMed PMID: 26910179
PubMedCentral PMCID: PMC4874862
Virological failure in patients with HIV-1 subtype C receiving antiretroviral therapy: an analysis of a prospective national cohort in Sweden
Analysis of clinical data reveals that patients with HIV-1C may have an increased risk of virological failure.
Haggblom A, Svedhem V, Singh K, Sonnerborg A & Neogi U. Lancet HIV 3, e166-174 (2016).
Pubmed PMID: 27036992

HIV-1 integrase binds the viral RNA genome and is essential during virion morphogenesis
A new role of integrase in formation of infectious HIV-1 particles is discovered.
PubMed PMID: 27565348
PubMedCenter PMCID: PMC5003418

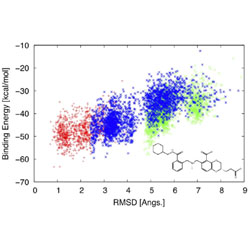
Binding energy distribution analysis method: Hamiltonian replica exchange with torsional flattening for binding mode prediction and binding free energy estimation
A new method for broadening the reach of a computational search provides better estimates of an inhibitor’s effectiveness.
PubMed PMID: 27070865
PubMedCentral PMCID: PMC4862910
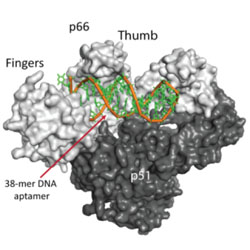
Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer
A DNA aptamer is used to determine the structure of reverse transcriptase without the need for crosslinking or a Fab.
Miller MT, Tuske S, Das K, DeStefano JJ & Arnold E. Prot. Sci. 25, 46-55 (2016).
PubMedCentral PMCID: 4815302
Factors influencing the efficacy of rilpivirine in HIV-1 subtype C in low- and middle-income countries
The development of resistance limits the use of rilpivirine as a first-line drug in HIV-1C-dominated epidemics.
PubMed PMID: 26518047
PubMedCentral PMCID: PMC4710214
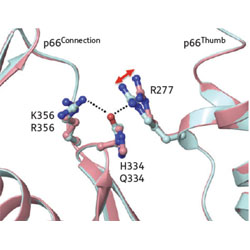
A new class of allosteric HIV-1 integrase inhibitors identified by crystallographic fragment screening of the catalytic core domain
HIVE researchers have discovered a new lead for design of integrase inhibitors, which shows improved action against resitant mutants
PubMed PMID: 27645997
PMCID: PMC5095411

Structural basis of HIV inhibition by translocation-defective RT inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA)
HIVE researchers reveal the mechanism of action of the most potent nucleoside analog inhibitor of HIV reverse transcriptase.
PubMed PMID: 27489345
PubMed Central PMCID: PMC4995989
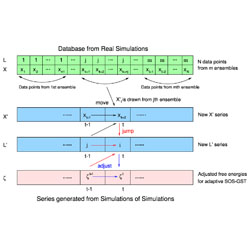
Locally weighted histogram analysis and stochastic solution for large-scale multi-state free energy estimation
A fast and accurate new method for estimating binding free energies is reported.
Tan Z, Xia J, Zhang BW & Levy RM J. Chem. Phys. 144, 034107 (2016)
PubMed PMID: 26801020
PubMedCentral PMCID: PMC4729400
Role of cysteines in stabilizing the randomized receptor binding domains within feline leukenia virus envelope proteins
Envelope protein is retargeted by addition of new cysteine amino acids.
Valdivieso-Torres L, Sarangi A, Whidby J, Marcotrigiano J & Roth MJ. J Virol 90, 2971-2980 (2016)
PubMed Central PMCID: PMC4810629

Chemoselective synthesis of polysubstituted pyridines from hetroaryl fluorosulfates
A selective new method is reported for creating inhibitors with polysubstituted pyridines
Zhang E, Tang J, Li S, Wu P, Moses JE & Sharpless KB Chem. Eur. J. 22, 5692-5697 (2106)
PubMed PMID: 26990693
PubMedCentral PMCID: PMC4929982

