HIV LIFE CYCLE
HIV Life Cycle (2015, David S. Goodsell)
These illustrations integrate information from structural biology, electron microscopy, and biophysical studies, with goal of simulating a view of the macromolecular structure of HIV in its cellular environment. Each illustration captures the virus at one point in its life cycle, in a cross section that shows all macromolecules and membranes. Current efforts are extending these semiquantitative illustrations, using these diverse sources of information to specify 3D models of HIV and its interaction with host cells, for use in hypothesis generation and simulation.
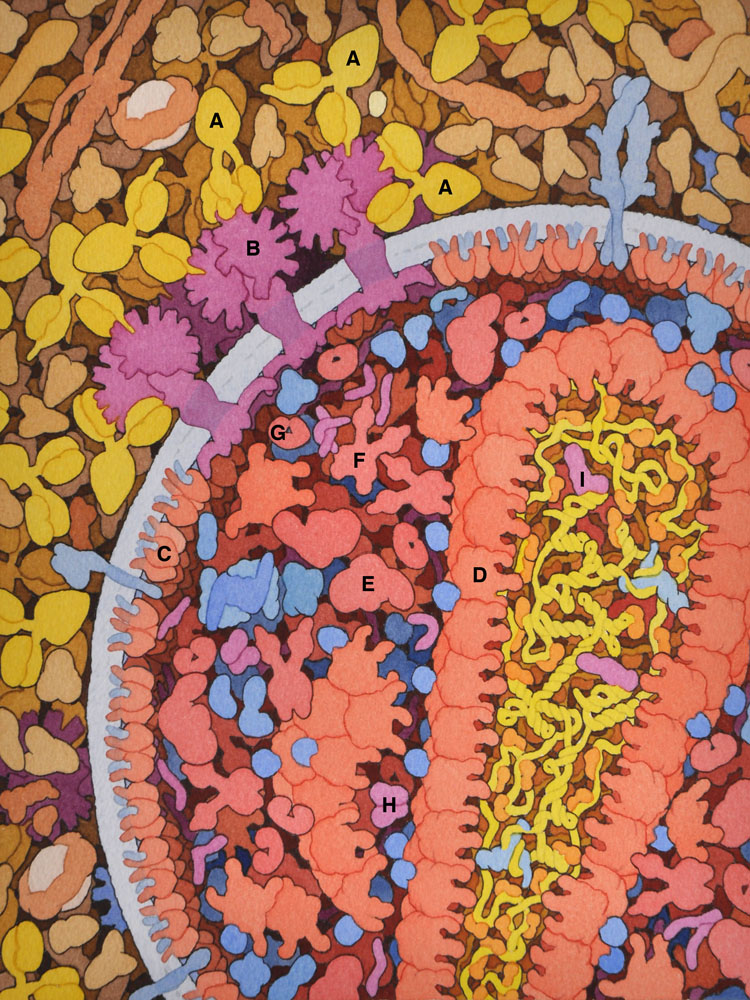
1 – HIV and Antibodies
In this cross section, HIV is shown at lower right, with viral proteins in red and magenta, and viral RNA in yellow. Blood plasma is shown at the top and left side. Several broadly-neutralizing antibodies (A) are binding to HIV envelope glycoprotein (B). Other viral proteins include matrix (C), capsid (D), reverse transcriptase (E), integrase (F), protease (G), Vif (H) and Tat (I).
2 – HIV Attachment
In this cross section, HIV is shown at the top and a target cell is shown at the bottom in blues. HIV envelope protein (A) has bound to the receptor CD4 (B) and then to coreceptor CCR5 (C), causing a change in conformation that inserts fusion peptides into the cellular membrane.
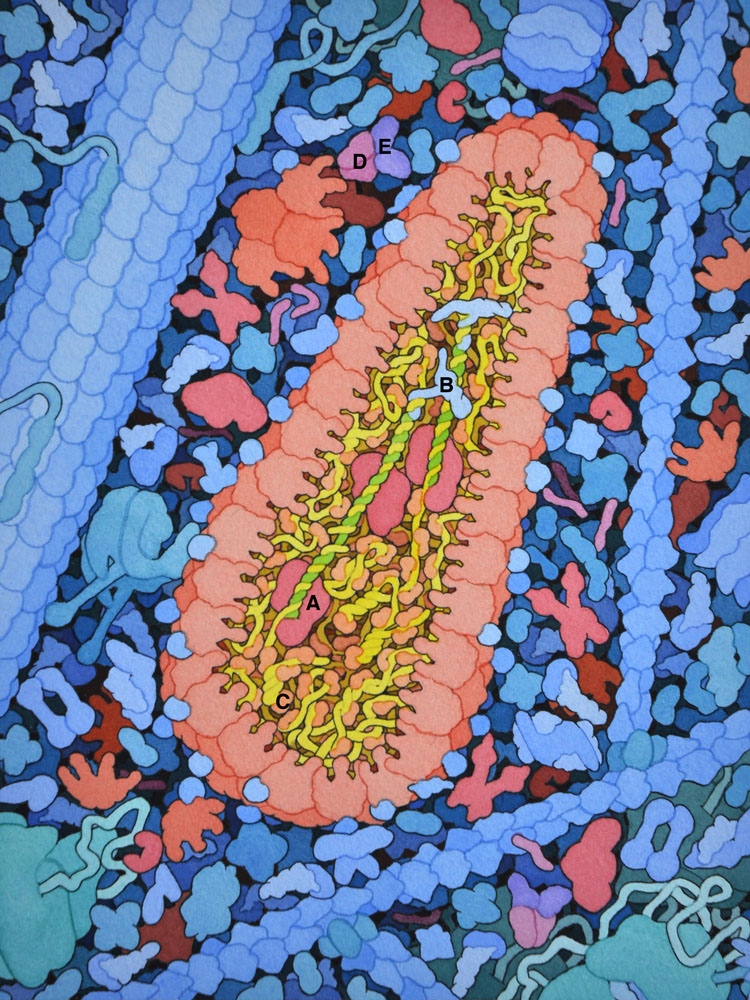
3 – HIV Reverse Transcription and Nucleocapsid
After the capsid has entered the cell, reverse transcriptase (A) creates a DNA copy (green) of the HIV RNA genome (yellow), using a cellular transfer RNA (B) as primer. HIV nucleocapsid protein (C) acts as a chaperone to unfold the RNA secondary structure. The ribonuclease activity of RT removes the viral RNA after the DNA strand is created. Interaction of HIV Vif (D) with cellular APOBEC (E) is also shown.
4 – HIV Integration
Uncoating of the viral capsid (shown at the top) and interaction with nuclear pore proteins such as Nup358 (A) releases the viral DNA (B). The DNA enters the nucleus through the nuclear pore (shown in purple) and is spliced into the cellular genome by the enzyme HIV integrase (C). Cellular protein LEDGF (D) is important for localization of the site of integration at DNA in nucleosomes (E).
5 – HIV Transcription and Tat Protein
HIV Tat protein (A), bound to the TAR RNA stem-loop structure, binds to the P-TEFb complex (B), activating transcriptional elongation by RNA polymerase (C). The illustration also shows HIV Rev (D) bound to the Rev-response element and CRM1 (E), a cellular protein involved in transport through the nuclear pore.
6 – HIV Translation
The HIV gag polyprotein (A, shown in red) is translated from the HIV RNA genome (in yellow) by cellular ribosomes (B). A stem-loop structure in the genome (C) induces a frame shift roughly 5% of the time, producing the longer gag-pol protein (D).
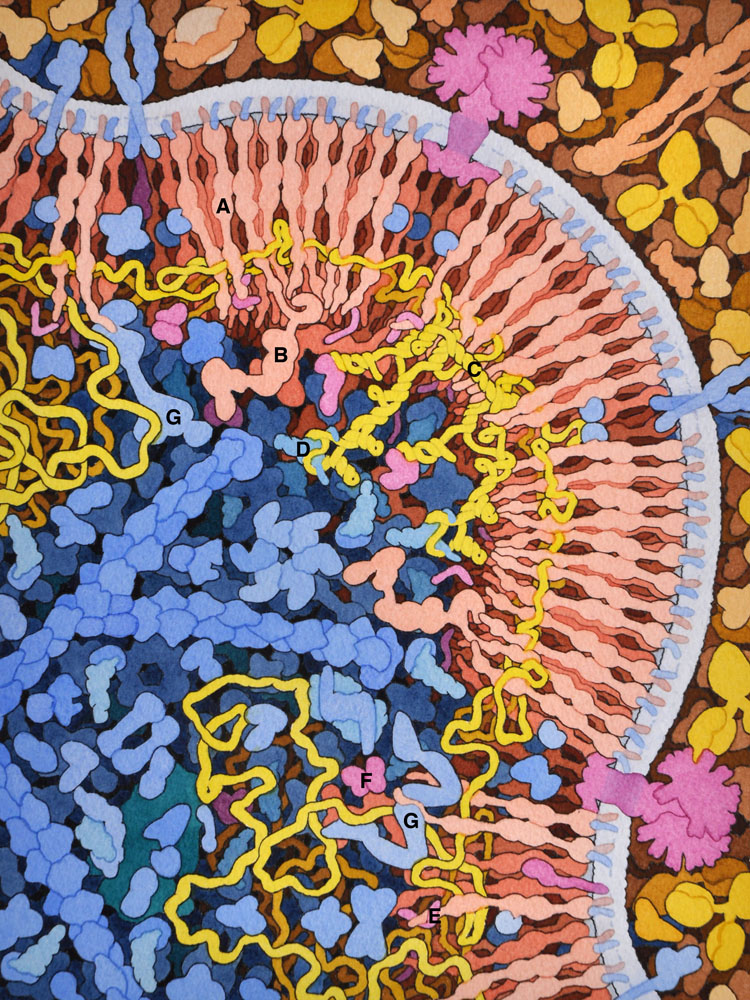
7 – HIV Budding
HIV gag protein (A) and gag-pol (B) form arrays on the cell surface, capturing two copies of HIV genome (in yellow), which dimerize through a specific sequence (C) and bind to a cellular transfer RNA (D) that will act as primer for reverse transcription. Viral proteins Vpr (E) and Vif (F) are also incorporated. Several cellular proteins of the ESCRT system (G) are involved in the process of budding.
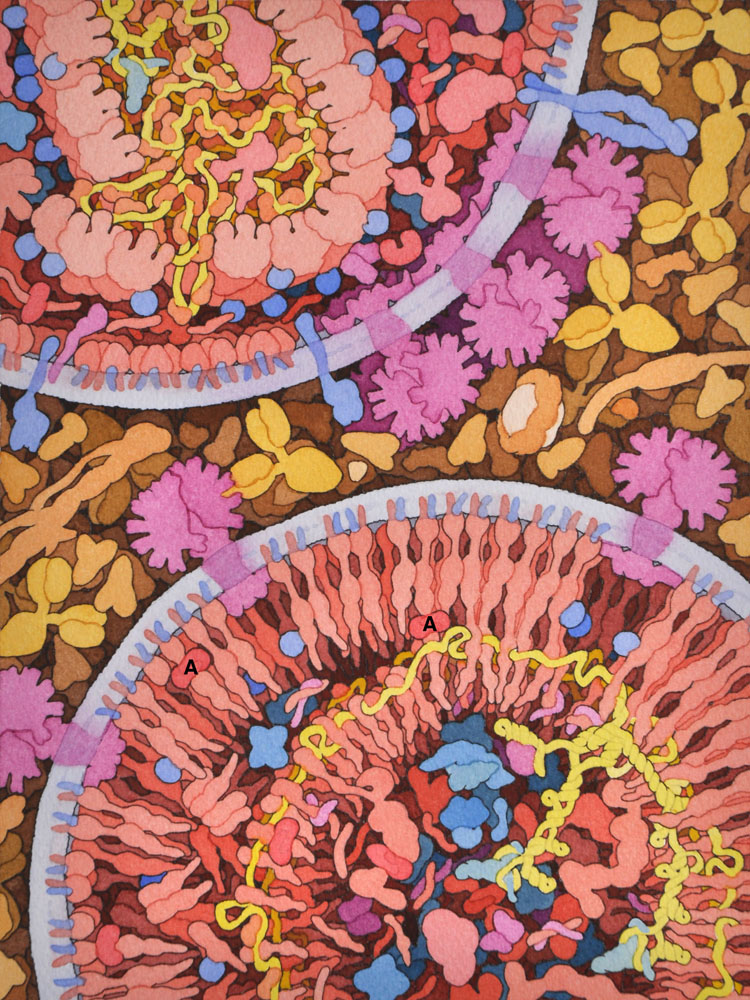
8 – HIV Maturation
This illustration shows an immature viron in the process of maturation at bottom right and a nearly-mature virion at upper left. HIV protease (A) is cleaving the gag and gag-pol proteins into functional proteins.