B-HIVE Structure Highlight: Neutral Sphingomyelinase 2
The genome of HIV includes instructions for building 15 proteins: structural proteins, a few enzymes, and accessory proteins that help seize control of infected cells. However, HIV also hijacks numerous cellular proteins to perform most of the tasks that it needs to produce new viruses. These include ribosomes and polymerases that manage production of viral proteins and many proteins involved in delivering everything to the right places during production of new viruses. B-HIVE researchers have recently discovered a new connection between a cellular protein, neutral sphingomyelinase 2, and key steps in the viral life cycle.
Tuning membrane composition
Neutral sphingomyelinase 2, shown here using a computed structural model from AlphaFold2, helps manage the complex composition of cellular membranes. It clips a small charged group off of the lipid sphingomyelin, producing ceramide. This small change can have huge consequences for the membrane. Ceramides, when they are added to one side of a lipid bilayer, can induce curvature in the membrane and create membranes that are less fluid. These studies have shown that HIV recruits neutral sphingomyelinase 2 to sites on infected cell membranes, to help create the proper membrane shape and consistency for new viruses as they bud from the surface.
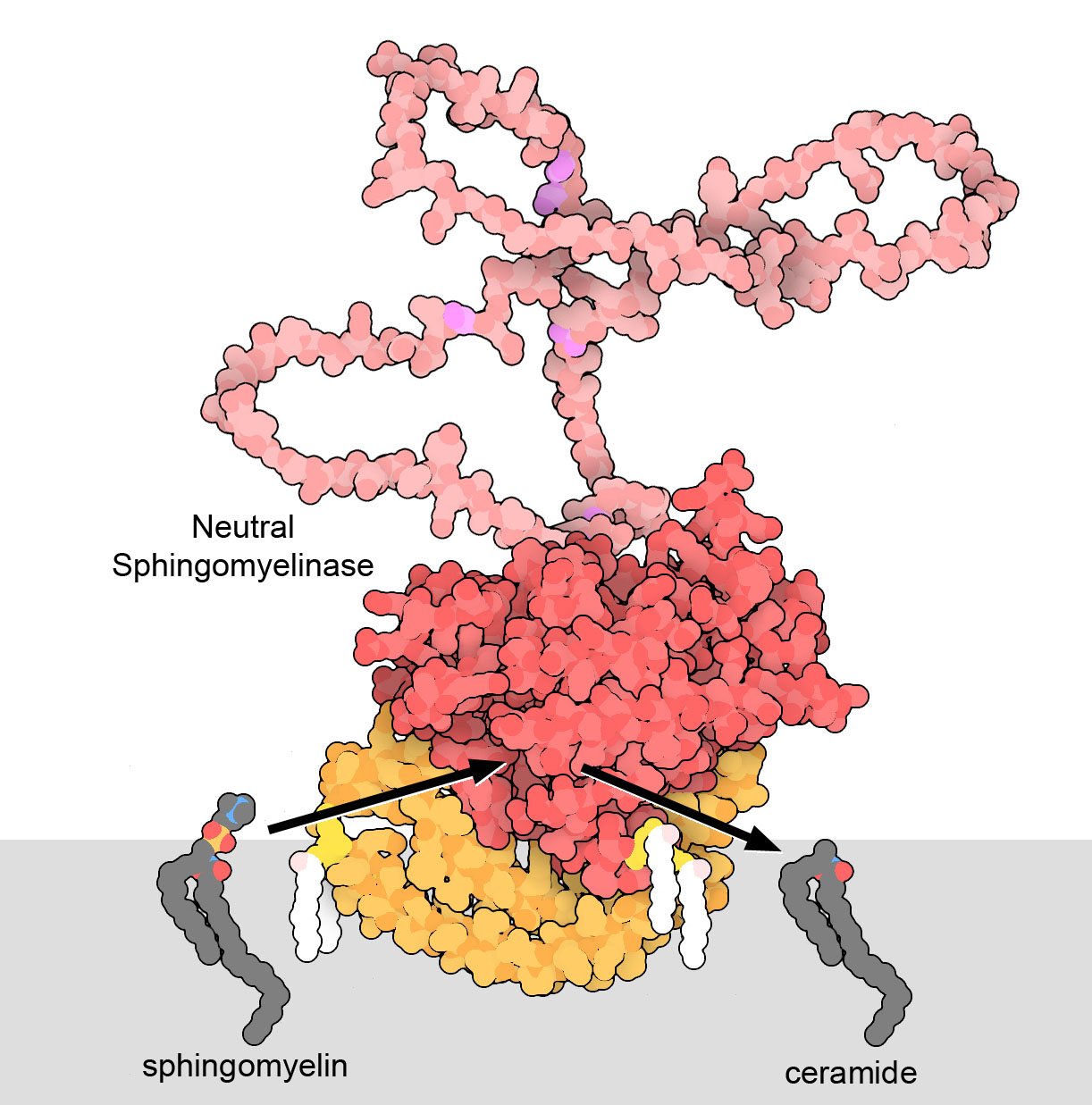
Neutral sphingomyelinase 2 includes several functional parts. The core of the protein (shown in red) is an enzyme that performs the lipid cleavage reaction. One end of the protein forms a membrane-binding domain (orange) that tethers the protein to the inner surface of the cell membrane, and includes several attached lipids (white). A long disordered loop (pink) extends into the cytoplasm and includes many serine amino acids (magenta) that can be phosphorylated to regulate the protein. The computed structure model was taken from PDB ID AF_AFQ9NY59F1.
What this study tells us about HIV-1
As part of this study, B-HIVE researchers found that inhibitors of neutral sphingomyelinase 2 block the production of infectious viruses. Looking closely, they found that deformed viruses are formed, with odd “blebs” on the surface and improper internal structures. Without the assistance of neutral sphingomyelinase 2, the virions were not able to mature to form the distinctive cone-shaped capsid of infectious HIV. These compelling observations reveal that neutral sphingomyelinase 2 may be an attractive target for development of new therapies to fight HIV-1 disease. As proof-of-principle, a new structurally novel inhibitor of neutral sphingomyelinase 2 was tested in humanized mice. In these mice, which have immune systems similar to the human immune system, the inhibitor blocked the production of viruses but also killed cells infected with HIV. This property has a highly beneficial consequence. The antiviral drug combinations in use today block replication of the virus, but HIV replication rapidly rebounds when the drugs are discontinued. In contrast, most of the humanized mice treated with the new inhibitor showed no rebound of viral replication after treatment was stopped.
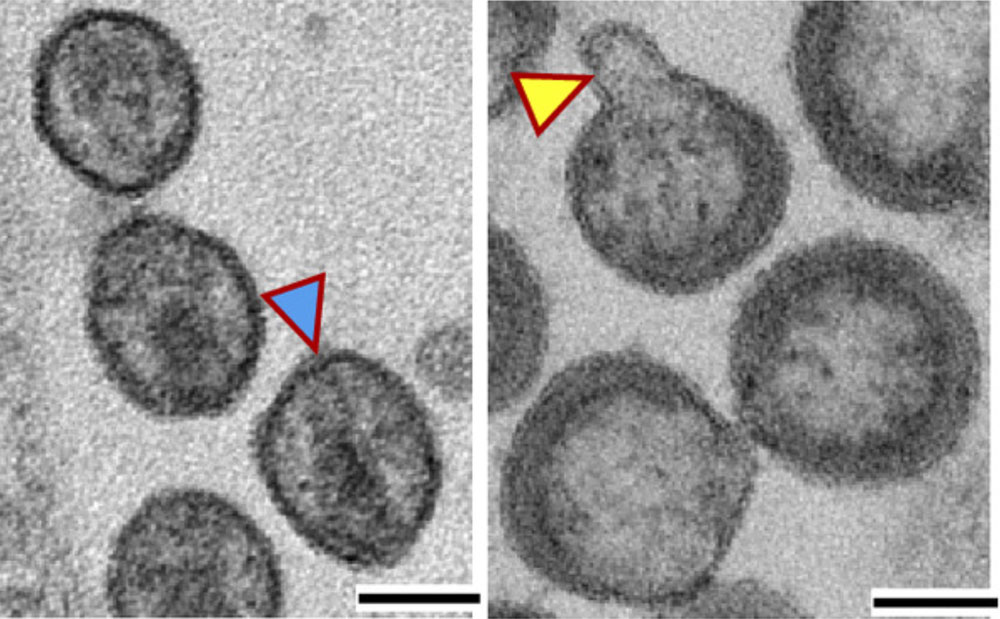
Electron micrographs of (left, blue arrow) a normal HIV virion and (right, yellow arrow) a distorted HIV virion produced when neutral sphingomyelinase is blocked. The scale bars represent 100 nanometers.
Meet the Researchers

Abdul A. Waheed
How did you get interested in science?
My journey into science was shaped by a combination of inspiring factors and academic interests. One of the key influences was an enthusiastic science teacher in high school who provoked my curiosity with engaging lessons and hands-on experiments. I developed a keen interest in chemistry, particularly organic chemistry, which acted as a driving force to understand the complex molecular structures that govern life’s processes. This interest in molecules and their interactions laid a strong foundation for my later exploration of biological systems at the molecular level.
Biomembranes have emerged as an appealing focal point within biological science, these barriers that define the boundaries of a cell and its organelles play a key role in cellular processes. The curiosity of how molecules cross these lipid bilayers fascinated me and prompted me to develop a deeper interest in understanding their biophysical and functional properties. My Ph.D. thesis was focused on understanding the mysteries of membrane dynamics and their implications in cellular function. During postdoctoral studies, my attention focused on lipid rafts – microdomains within biomembranes that have significant implications in cell signaling. Exploring their role in various processes, including virus assembly, opened up a new avenue that bridged membrane biology and virology.
In essence, my journey into biological science was a symphony of inspiration from mentors, a passion for molecular interactions, and a continuous commitment to understand the fascinating complexities underlying living organisms.
Tell us about the lab where you did this work.
I conducted this research at the laboratory of Dr. Eric Freed at the National Cancer Institute (NCI) in Frederick, Maryland. Dr. Freed is a well-known researcher in the field of molecular virology, with an emphasis on the late stages of the HIV-1 replication cycle. His research focuses on HIV-1 Gag trafficking, virus assembly and release, maturation, and drug resistance. The NCI-Frederick campus is renowned for its state-of-the-art research facilities and its contributions to advancing our understanding of cancer, infectious diseases, and related fields.
Around two decades ago, I started an exciting chapter of my scientific journey by joining Dr. Freed’s lab. My expertise in the field of lipid rafts during my prior postdoctoral work facilitated this transition. This move to Dr. Freed’s lab marked a significant advancement in my career, aligning perfectly with my passionate interest in lipid rafts and their functional significance. Dr. Freed’s lab, renowned for its pioneering research in the late stages of the replication cycle, provided an ideal platform to explore the connections between lipid rafts and HIV-1 assembly. I was able to merge my expertise in lipid rafts with the lab’s primary focus on retroviral assembly. Over time, my research shifted towards investigating host-restriction factors in the HIV-1 life cycle, such as tetherin, TIM-1, SERINC, PSGL-1, and MARCH proteins.
The invaluable guidance and support of Dr. Freed encouraged a sense of confidence that empowered me to undertake ambitious projects. With his support, I embarked on scientific endeavors that stretched my limits, expanded my horizons, and significantly contributed to my scientific and personal growth. The bonds formed with my colleagues in Dr. Freed’s lab transformed it into a second family for me. This bond we shared was instrumental in making the lab a place for vibrant scientific exploration and personal development.
What were the biggest challenges with this study?
One of the major challenges in this study is understanding how disruption of nSMase2, either through siRNA-mediated knockdown or treating virus-producing cells with selective inhibitors like PDDC or DPTIP impairs HIV-1 Gag and GagPol processing, resulting in a profound impairment in particle maturation and infectivity. We hypothesize that alterations in the lipid composition, particularly reduced ceramide levels due to nSMase2 inhibition, impact viral membrane properties. These changes in the biophysical properties of the viral membrane, in turn, hinder GagPol dimerization, a critical step for cleaving the GagPol and Gag precursor proteins into their individual domains, allowing the virus to mature. To investigate these biophysical changes, we use fluorescence probes such as Laurdan to assess viral membrane fluidity. We employ FRET between fluorescently tagged GagPol proteins to investigate GagPol dimerization under nSMase2 disruption.
To further understand the mechanism of action of nSMase2 inhibitor, we selected PDDC-resistant HIV-1, and mutations that confer resistance to PDDC were mapped to the MA and CA domains of Gag. However, these mutants exhibited only low-level resistance to PDDC. Our objective is to develop high-level resistance to PDDC and identify mutations that confer resistance. These studies will shed some light on the mechanism of action of this nSMase2 inhibitor.
We discovered that nSMase2 disruption significantly impairs the maturation and infectivity of other primate lentiviruses, such as HIV-2 and simian immunodeficiency virus. However, it has a moderate or no effect on non-primate lentiviruses like equine infectious anemia virus and feline immunodeficiency virus, and has no effect on the gammaretrovirus murine leukemia virus (MLV) and alpharetrovirus Rous sarcoma virus. To explore PDDC sensitivity, we examined Gag chimeras from sensitive (HIV-1) and insensitive (MLV) viruses to map determinants of nSMase2 disruption sensitivity. Intriguingly, our findings suggest that HIV-1 Pol is the target of nSMase2 disruption. Currently, we are examining determinants within HIV-1 Pol for sensitivity to nSMase2 disruption.
What are you working on now?
Continuing my research, my focus is on resolving the mechanism behind Gag and GagPol processing defects during nSMase2 disruption. This involves investigating viral membrane biophysical properties, selecting high-level PDDC resistance and identifying mutations, and pinpointing molecular determinants in HIV-1 GagPol that are sensitive to nSMase2 disruption.

Seung-Wan Yoo
How did you get interested in science?
When I first chose my major, Genetic Engineering, in college, I wasn’t particularly interested in science. Like many students my age, I was more focused on enjoying the new college experience after high school. However, things changed when I had to fulfill my mandatory service period in the army for two years. During my time in the army, I started thinking more seriously about my life and developed an interest in life sciences, especially neuroscience. This newfound fascination led me to decide to pursue a career in neuroscience. After completing my mandatory service and returning to college, I dedicated a significant amount of time to studying my major. Eventually, I made the decision to enter graduate school, where I earned my Master’s degree in the study of protein methylation and acetylation in the field of neurobiology. My journey in the field of neuroscience continued during my Ph.D., where I focused on stem cell therapy for stroke. Subsequently, I continued my academic journey by joining Dr. Ronald Schnaar’s lab at Johns Hopkins University as a postdoctoral fellow from 2011 to 2013. During this time, I delved into understanding the roles of sialoglycans in brain structure and function using genetic knockout mice. In 2013, I became a part of Dr. Norman Haughey’s lab at Johns Hopkins University, where I am currently located. My research focus shifted to understanding how changes in sphingolipid metabolism are connected to the development of HIV-related complications in the central nervous system (CNS).
Tell us about the lab where you did this work.
Our laboratory, led by Dr. Norman Haughey, is situated within the Johns Hopkins University School of Medicine in Baltimore, MD. Our research focus revolves around a disease-centered research program that delves into fundamental neurobiology and clinical neurology. Our primary research interests encompass: (1) Biomarker Identification: We’re engaged in diverse research projects aimed at identifying biomarkers for neurodegenerative conditions such as HIV-Associated Neurocognitive Disorders, Multiple Sclerosis, and Alzheimer’s disease. This involves analyzing metabolic profiles in blood and cerebral spinal fluid samples from ongoing clinical studies. We employ a range of advanced techniques, including biochemical analyses, mass spectrometry, and bioinformatics. These biomarkers can then be used in the diagnosis of disease, as prognostic indicators to predict disease trajectory, or as surrogate markers to track the effectiveness of disease modifying interventions. (2) Lipid-Protein Interactions: We’re dedicated to unraveling the intricate interplay between lipid components present in neuronal and glial membranes and proteins. This interaction governs critical processes like signal transduction associated with differentiation, motility, inflammatory signaling, survival, and neuronal excitability. (3) Extracellular Vesicles (Exosomes): Our research delves into the influence of extracellular vesicles, particularly exosomes released by brain resident cells. We explore how these vesicles regulate neuronal excitability, neural network activity, and immune responses in peripheral regions following damage or infections within the central nervous system.
Our multifaceted research efforts encompass a wide spectrum of neurobiology, from molecular interactions to clinical applications, with the ultimate aim of advancing our understanding of neurodegenerative diseases and developing strategies for their diagnosis, treatment, and management.
What were the biggest challenges for this study?
One of the most significant challenges I faced during this study was the creation of humanized mice, an experimentation that we had not experienced before. This process was met with various obstacles, including a notably high mortality rate among the mice post-humanization, insufficient levels of humanization achieved, and challenges with achieving effective HIV infection. Over a span of several rounds of trial and error, as well as valuable guidance from Larisa Pluektova at the University of Nebraska, we eventually managed to successfully create our initial set of eight humanized mice. This accomplishment came approximately two years after we began this endeavor. Another major challenge we encountered was assessing the efficacy of PDDC (an inhibitor of nSMase2) in HIV-infected humanized mice. While we ultimately utilized chow containing PDDC to treat the mice at a later stage of the study, our initial approach involved daily intraperitoneal injections of PDDC for several years. This administration continued even during the COVID-19 pandemic when access to the lab was restricted. After obtaining an essential personnel status, I could enter the lab, working for minimal durations while ensuring consistent dosing of PDDC to the HIV-infected mice.
What are you working on now?
I am continuing to unravel the complex mechanism by which nSMase2 exerts its regulatory influence on HIV-Gag processing and maturation. Employing a combination of techniques in molecular biology (functional domain mutation) and biochemistry (functional activity analysis), I will dissect the steps and interactions involved in this process. Furthermore, my research will attempt to improve the understanding of PDDC-induced cell death. Through a combination of experimental techniques, I will delve into the intricate network of signaling pathways and interactions that emerge in response to endolysosomal stress triggered by PDDC. By dissecting these crosstalk mechanisms, I aim to elucidate the specific ways in which this stress influences cell-death regulation. This investigation holds the potential to unveil novel insights into the complex interplay between endolysosomal stress and cell death pathways, contributing to a deeper understanding of therapeutic interventions targeting endolysosomal stress-induced cell death.
