B-HIVE Structure Highlight: 3D modeling for protein structure and function discovery
3D Modeling
If a picture is worth a thousand words, then an animated 3D model of a complex system, such as a virus or cell, is surely worth much, much more. Models depict what a virus or cell looks like in a biologically realistic context and provide insights into its structure and function.
Supported by a B-HIVE Collaborative Development Grant, Ludovic Autin recently established a lab at the Scripps Research Institute dedicated to modeling and visualizing how proteins and other macromolecules are organized within organelles and other biological compartments. His lab develops specialized software and uses molecular modeling to create integrative models that represent current knowledge of a given system, including HIV-1. His lab builds mesoscale models as realistic and biologically faithful as possible, integrating and leveraging cryo-electron tomography data to guide their construction.
CellPACK
One of Autin’s most significant accomplishments is the software suite CellPACK. This software builds integrative structural models ranging in scale from viruses to small organelles and even whole bacteria. Autin developed CellPACK with colleagues Graham Johnson and David Goodsell. Goodsell is well-known for his paintings of molecular-level biochemical processes.
Early efforts in CellPACK were limited to small viruses like HIV-1. A collaboration with Ivan Viola and Mathiew Le Muzic, however, led to a rendering engine capable of displaying, in real time, billions of atoms. This enabled an atomic-level model of viruses like HIV-1.
CellPACK has also created full atomic-level models of cells of the bacterium Mycoplasma genitalium, including around 27,000 proteins. Autin and collaborators, including Rob Dick of B-HIVE, are working on modeling other viruses like the Nipah virus, influenza, and coronaviruses. Other collaborators are using CellPACK to model specific organelles, such as mitochondria.
How this lab is advancing HIV research
One of the lab’s current focuses is supporting machine learning methods to analyze cryo-electron tomography data and advance research into the structure of HIV-1. A key application of their 3D modeling work is to support processing of tomography data. At the moment, researchers still manually label regions of the tomogram to identify compartments and protein particles. While new tools are beginning to automate parts of this task, it remains a substantial and tedious amount of work. The Autin lab is exploring is whether “painting” a 3D protein model directly onto the tomogram could help accelerate the identification and reconstruction of cellular or viral structures.
With atomic-level models, Autin can also generate synthetic electron microscopy datasets. Currently, machine learning models designed to analyze electron microscopy images don’t have enough data for thorough and robust training. Synthetic datasets, however, can rapidly and significantly expand the amount of data available for machine learning training. If machine learning models can gain proficiency and efficiency in tasks like segmenting experimental cryo-electron tomography data, then research into the structure and function of cells and viruses like HIV-1 can, hopefully, accelerate.
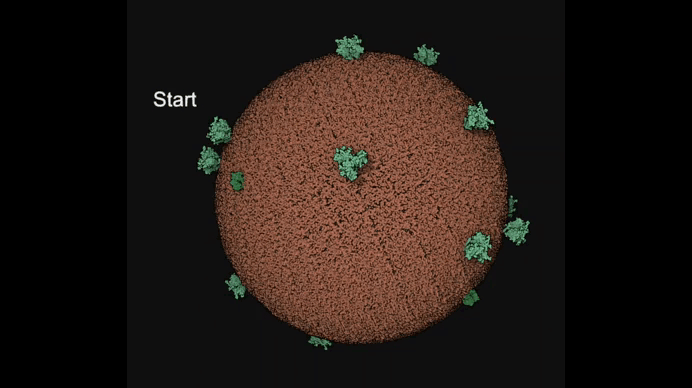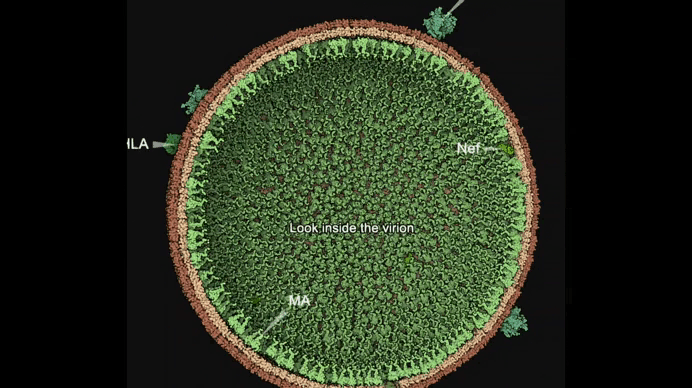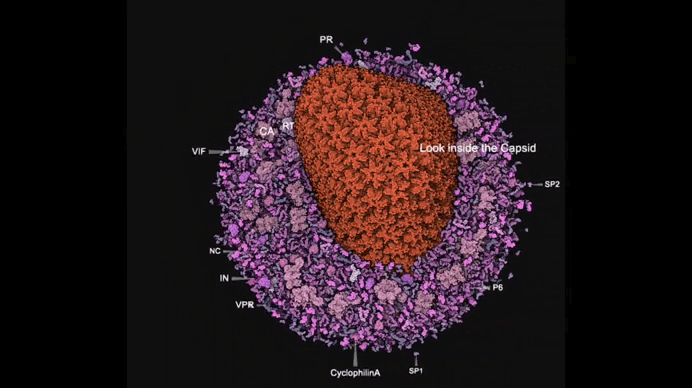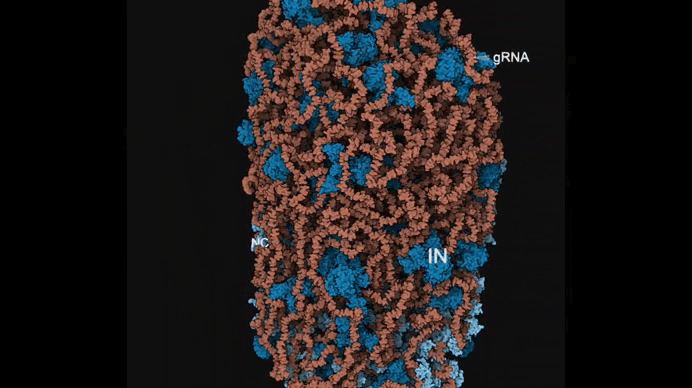

Autin and colleagues use cellPACKgpu to build integrative models of the mature HIV (top), with a focus on the Capsid fullerene cone (bottom). Learn more at https://autinlab.org
Meet the Researcher
How did you get interested in science?
My interest in science began in high school, where I was captivated by biology—especially the complexity of hormonal pathways and the principle of feedback regulation. What struck me most was the realization that such intricate regulatory systems can actually emerge from seemingly simple molecular interactions at the atomic level.
In college, I majored in biology. One year, I got the opportunity to work at a veterinary school. I was working on an application to explain how a parasite attacks animals. I was in a unit where people were starting to work with 3D. Someone introduced me to 3ds Max, and I was just blown away by what you could do with it. It was really fun. I played around with it, making little movies with spheres and cones.
After that, I joined the lab of professor Jacques Chomilier specializing in predicting the structure of protein loops. That’s when I discovered that I could create graphical representations for proteins and import them in 3ds Max. And that was a tipping point. We can do really cool stuff with protein structure. The same tools used in artistic 3D movie production could also be applied to biology. This opened the door for me to visualize and interact with proteins, sparking a passion for molecular graphics and driving my motivation to keep pushing the boundaries of what we can model and represent. After that, I was fortunate to join Bruno Villoutreix’s lab in Paris for my PhD, where I developed, using docking methods, models of protein–protein interactions involved in blood coagulation, while also nurturing my curiosity for 3ds Max and molecular illustration. I actually made extensive use of 3ds Max for the illustrations in my thesis. From there, it felt completely natural to join Art Olson’s Molecular Graphics Laboratory at Scripps Research as a postdoctoral fellow, bringing together all these interests at the intersection of computation, visualization, and biology.
Tell us about the lab where you did this work.
Today, I still work at The Scripps Research Institute, where I’ve recently taken over David Goodsell’s lab. My work follows a long tradition of blending art and science, shaped by my close collaborations with David and Art. Our space reflects that heritage—we keep old 8mm projectors and 3D-printed molecular models around, which give the lab an artistic atmosphere. I strive to carry forward this combination of creativity and science through ongoing efforts in molecular graphics and in outreach development. For example, we use Meta Quest headsets with visitors to immerse them in the intricate world of viruses and to illustrate the fascinating quasi-symmetry principles that govern their structure.
What are the biggest challenges you’re facing?
One of the biggest challenges today is keeping pace with the rapid advances in machine learning across so many areas of research. At times it can feel overwhelming.
I can give you a couple of examples. A year ago, vibe coding was not a thing. Any code writing with AI tools was not usable. As of today, it is. So, if tomorrow I have an idea about a project, I can kickstart the project, because AI will be able to generate code that can run right away.
But in terms of processing electron microscope data, it’s even more tricky. Even though there are new papers coming out frequently about new methods to segment or automatize the segmentation of data, they are either really hard to use or come from different fields—like an algorithm that works really well and segments exactly where you clicked, but it’s for a 2D image. It doesn’t work for volumetric data.
Still, I believe there remains an important place for handcrafted, hypothesis-driven work, and I see these new methods not as a replacement, but as powerful tools at our disposal to help us tackle the mysteries of the inner cell.
What are you working on now?
We are focusing on the assembly of the HIV capsid. While it has been widely studied and we know a great deal about it—for example, that it forms a fullerene cone shape built from pentamers and hexamers of the capsid protein—we still lack efficient ways to remodel it. Segmentation of this structure is still largely done manually, which is both slow and limiting. Our goal is to automate the generation and segmentation of the capsid directly from experimental data such as cryo-electron tomography images. By doing this, we will be able to provide paired datasets—synthetic and experimental—that can serve as a test bed for training deep-learning models. Ultimately, this will help us move toward more automated and scalable understanding of viral structures.

